-
Type:
Bug
-
Status: Closed
-
Priority:
Major
-
Resolution: Cannot Reproduce
-
Affects Version/s: 39, 38.1, 40, 41, 40.1, 42
-
Fix Version/s: None
-
Component/s: Basic-Nucl, seq
-
Tests Type:Functional/Unit
-
Affect Type:Userdefined
Reproduced in version 38.1, but the bug may have existed before.
- Open _common_data/pcr_primer_design/gfp.fa.
- Open the OP "In Silico PCR" tab.
- Fill in:
- Forward primer: GAACTATATTTTTCAATGGGCACAAATTTTCTGTC;
- Reverse primer: TACTCATTTCCTCTTCTTGAAAAGTGACCTCAACAGGGTTAAGAACAACTTAATCTACCACTACAATTACCCGTGTTTAAAAGACAGTCACCTCTCCCACTTCCACTACGTTGTATGCCTTTTGAATGGGAATTTAAATAAACGTGATGACCTTTTGATGGACAAGGTACCGGTTGTGAACAGTGATGAAAGCCAAT;
- 3' perfect match: 0.
- Repeat the following steps many times (~20 or more):
- Click "Show primers details". The "Primer Details" dialog appears;
- Enlarge the dialog to see the heterodimer characteristics (Delta G and Base Pairs);
- Expected: the hetero-dimer and its characteristics are always the same across runs, e. g. "Delta G=-7.4, Base Pairs=5",
Current state: hetero-dimers may differ, for example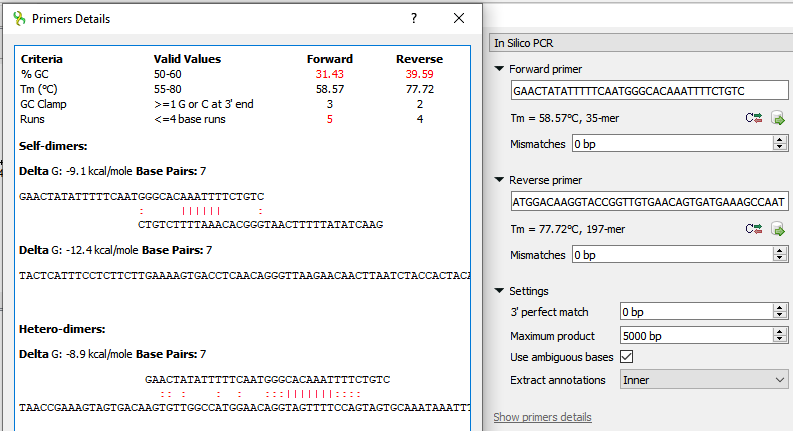
- Close the dialog (you can use Alt+F4) and repeat the steps until the bug appears.
Perhaps repetitive steps need to be done quickly.