- Open human_T1.fa.
- Open "In silico pcr" tab
- Forward primer - GTGTAAAATATGCAGAAGAGCATCTAACA
Reverse primer - GCTCAAAAACCCAATTCTAAGGGG - Click "Show primer details".
- Look at the first "Self-dimer".
Current:
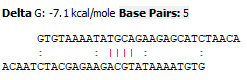
Important, that Base Pair: 5
Expected: (the same dimer, but from OligoAnalyzer)

Base Pairs: 4
PS, I am pretty sure, that I have already faced and fixed this in Pcr Primer Design, but we did not merge this branch. Check this - I am sure, this is the working fix.
- blocks
-
UGENE-7918 Create new Primer3 dialog "Presets" feature and add a new preset called RPA (Recombinase polymerase amplification)
-
- Closed
-